Formaldehyde is known to be a human carcinogen which do a lot of harm to human beings, especially whose house is newly decorated. And to make matters worse, there are no effective and non-secondary-pollution method to eliminate indoor formaldehyde. To solve this problem, we developed a method of detection and removal of indoor formaldehyde.
Using synthetic biology, we developed a method of detection and removal of indoor formaldehyde, which can solve this problem without secondary-pollution.
Background: About Formaldehyde
Properties
Formaldehyde is the simplest aldehyde. It is a highly reactive gas and is formed by oxidation or incomplete combustion of hydrocarbons. Formaldehyde exists at room temperature as a nearly colorless gas with a pungent, suffocating odor. In its pure state, formaldehyde is not easily handled, because it is extremely reactive and polymerizes readily. The primary form of formaldehyde in dilute aqueous solutions is its monomeric hydrate, methylene glycol (methanediol) and the primary forms in concentrated solutions are oligomers and polymers of polyoxymethylene glycols.[1]
The physical and chemical properties of formaldehyde are summarized in Table 1.
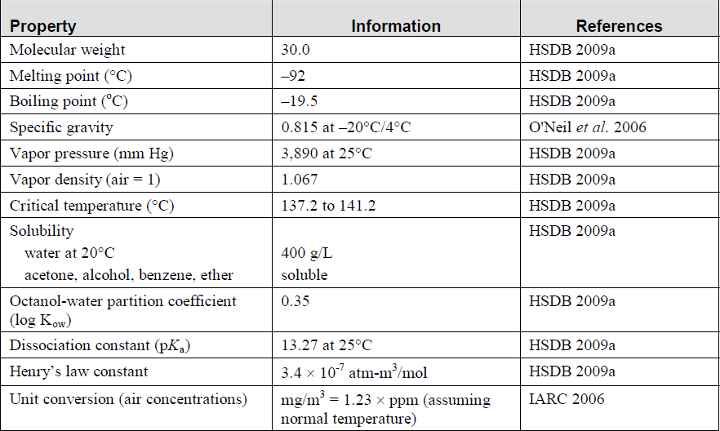
Table 1. Physical and chemical properties of formaldehyde
Formaldehyde is an important precursor to many other materials and chemical compounds. In view of its widespread use, toxicity and volatility, exposure to formaldehyde is a significant consideration for human health. In 2011, the US National Toxicology Program described formaldehyde as "known to be a human carcinogen".[2]
Applications
3.1 Construction and Decorative Products
Since the invention of the particle board in Germany in the 1940s, the use of formaldehyde in glues and resins has revolutionised the construction industry. Formaldehyde-based resins allow wood chips, sawdust and even some recycled wood waste to be combined to create wood products that are durable, cost-effective and high performance alternatives to solid wood. Formaldehyde-based resins also provide better dimensional stability and mould resistance.
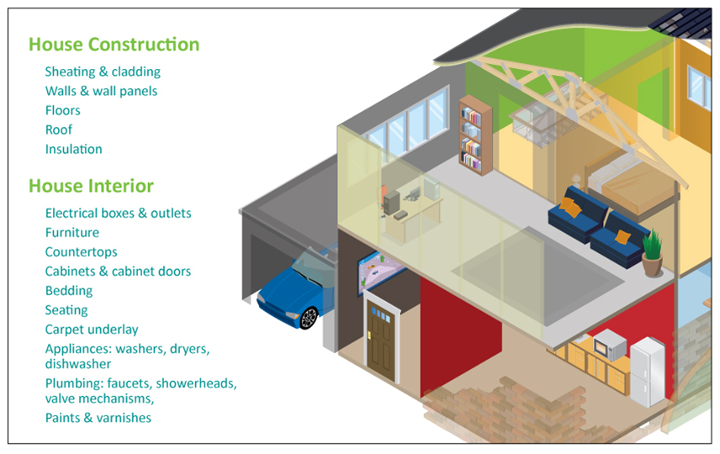
Figure 1: Formaldehyde in Construction and Decorative Products
Automotive Applications
Many 'under-the-hood components' such as fuel pumps, transmission parts and brake pads are produced with formaldehyde-based resins. Other formaldehyde based applications include the decorative laminates of car interiors, engine lubricants, vulcanized rubber tyres and lightweight polyurethane foams for automobile door insulation.

Figure 1: Formaldehyde in Construction and Decorative Products
Harm
Toxic effects
Formaldehyde is a highly reactive chemical that causes tissue irritation and damage on contact. Formaldehyde concentrations that have been associated with various toxic effects in humans show wide interindividual variation and are route dependent. Symptoms are rare at concentrations below 0.5 ppm; however, upper airway and eye irritation, changes in odor threshold, and neurophysiological effects (e.g., insomnia, memory loss, mood alterations, nausea, fatigue) have been reported at concentrations ≤ 0.1 ppm. The most commonly reported effects include eye, nose, throat, and skin irritation.
Carcinogenicity
Formaldehyde is known to be a human carcinogen based on sufficient evidence of carcinogenicity from studies in humans and supporting studies on mechanisms of carcinogenesis.
Epidemiological studies have demonstrated a causal relationship between exposure to formaldehyde and cancer in humans. Causality is indicated by consistent findings of increased risks of nasopharyngeal cancer, sinonasal cancer, and myeloid leukemia among individuals with higher measures of exposure to formaldehyde (exposure level or duration), which cannot be explained by chance, bias, or confounding.
Formaldehyde exposure is associated with multiple modes of action related to carcinogenicity, such as DNA reactivity, gene mutation, chromosomal breakage, aneuploidy, epigenetic effects (binding to lysine residues of histones), glutathione depletion, oxidative stress, and cytotoxicity-induced cellular proliferation (Lu et al. 2008, Guyton et al. 2009, NTP 2010). There is evidence for a genotoxic mode of action for formaldehyde-induced cancer; however, the mechanisms by which formaldehyde causes cancer are not completely understood and most likely involve several modes of action.[1]
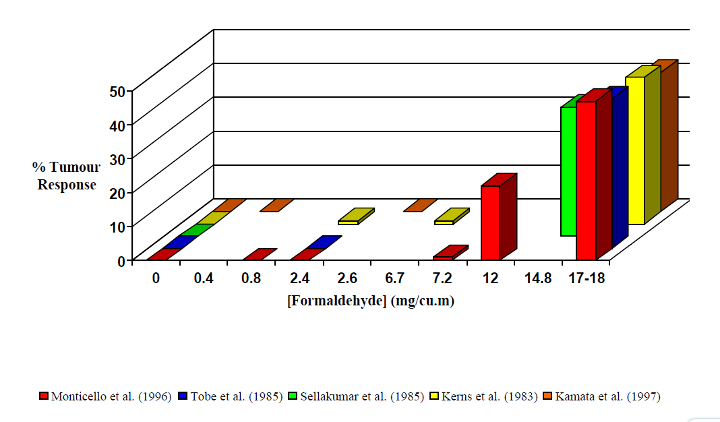
Figure 1: Formaldehyde in Construction and Decorative Products
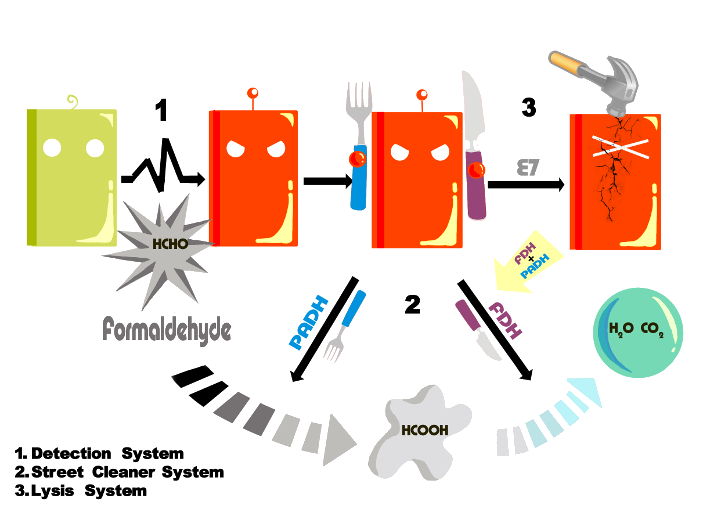
To detect and remove the formaldehyde, we design and construct three systems to build our Formaldehyde Terminator.
Coloration system
Since we would like to use formaldehyde as an input in our design,can we find or construct a biobrick sensing the presence of formaldehyde? In Bacillus subtilis 168, there is an operon called hxlAB operon.[1][2]

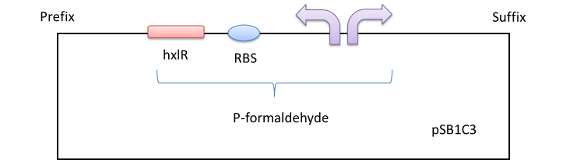
Figure 1 The formaldehyde promoter shown in the red column
This operon controls the expression of two key enzymes in the ribulose monophosphate pathway that are involved in formaldehyde fixation, 3-hexulose-6-phosphate synthase and 6-phospho-3-hexuloisomerase. [3] Expression of the hxlAB operon is induced by the presence of formaldehyde. HxlR protein can enhance the operon to some extent. The hxlR gene that encodes HxlR protein is located upstream the hxlAB operon.
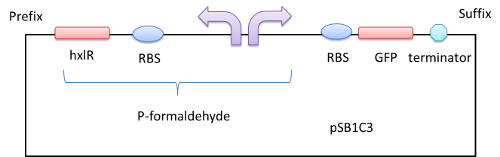
We get a part of the operon (which we name it P-formaldehyde,shown in a red column in Figure 1) by PCR , add a fluorescent protein or chromoprotein downstream and transform the plasmid(Figure2) into E.coli DH5a. In this way, we can know whether there is formaldehyde or not from the color change.
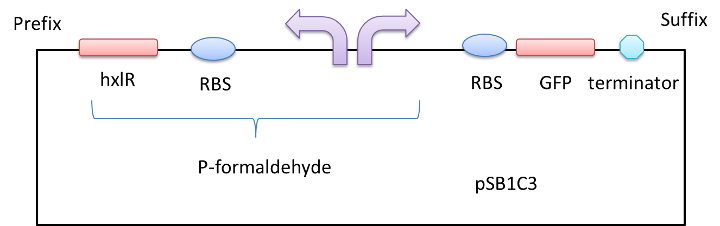
Figure 2 A GFP reporter is added downstream of the formaldehyde promoter
The street cleaner system
PADH, formaldehyde dehydrogenase from Pseudomonas putida is a formaldehyde dehydrogenase which can oxidize free formaldehyde to formate independent of GSH. [4] FDH, formate dehydrogenase from Candida boidinii can oxidize formate to carbon dioxide in the presence of NAD+.[5][6]
Figure 3 The mechanism of two enzymatic reactions
We would like to use this two enzymes to degrade formaldehyde into water and carbon dioxide. In our design,the expression of this two enzymes can be controlled by the P-formaldehyde so that the presence of formaldehyde can induce the expression of them. We can also use constitutive promoter to guarantee the expression level.
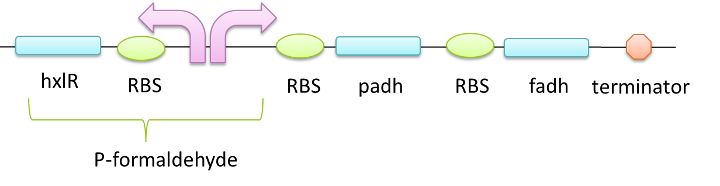
Figure 4 Two enzymes are added downstream of the formaldehyde promoter
Lysis system
Lysis system has two important roles in our design. One is about biosafety; the other is to help the enzymes come out of the bacteria so that they can work better in the environment.
We use Lux system, E7 protein and our P-formaldehyde to construct this system.
Lux system is a quorum sensing system , consisting of luxI, luxR and P-lux. luxI is a synthase for a small molecule called acyl-homoserine lactone (AHL). AHL can bind to the LuxR protein and then the composite can stimulate transcription from the right hand lux promoter P-lux.[7]
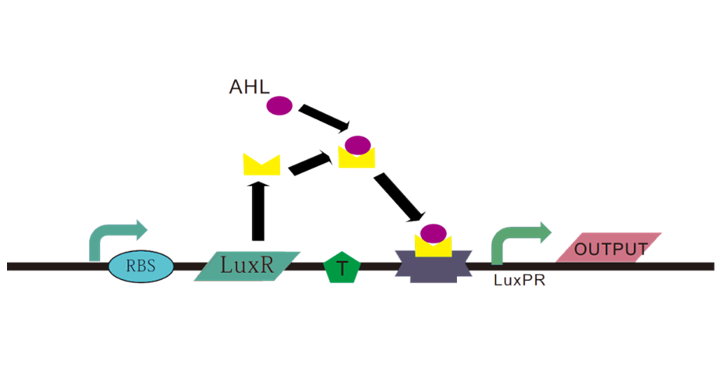
Figure 5 The mechanism of the lux system
The E7 lysis protein is a key component of the SOS response system in E.coli and functions to export bacteriocins into the extracelluar space under stressful environmental conditions. Besides, this protein only has 47 amino acids and thus can be easily utilized as an modular part in a constructed biobrick which helps the expressed foreign protein easier to be collected or take effect outside cell.[8][9] In this case, E7 lysis protein is used to release enzyme FDH and enzyme PADH to the extracellular space so that they can degrade formaldehyde in a more efficient way.We design this system as following:
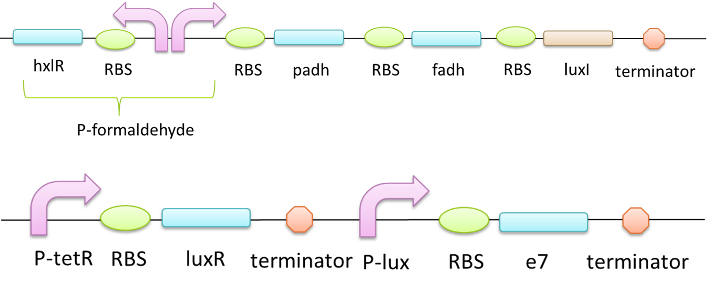
Figure 6 A system which can release two enzymes out of the cells when stimulated by formaldehyde
When the P-formaldehyde is activated, the bacteria will express FDH, PADH and LuxI protein. LuxI synthesis AHL and AHL binds to LuxR so that the P-lux is activated and the E7 protein is expressed. When E7 reaches a certain level, the bacteria lyse.
Application:
To make our detection and street cleaner systems more convenient and functional for others to use in both daily life and industry (this is also a requirement for a good iGEM project), we have made a device (see "A Device for Daily Life") and got a detailed plan about industrial application (see "Devices for Industrial Application").
A Device for Daily Life
We have made a convenient device(showed below) to hold our E.coli so that we can detect the formaldehyde gas easier. A electronic formaldehyde detector was added so that we can compare our detection system with it.

In our future work, we will combine this device with automatic recording machine. By connecting them, we can get data automatically and precisely. Then we will optimize it by diminishing its volume and embellishing its appearance so that it can be easier to use.
Devices for Industrial Application
Although there still remains a lot for our project to come to be used practically, we can hope that one day our dream can come true. As we know from the information above, our systems can be used separately; one way is that our Street Cleaner System can help to clean formaldehyde from industrial effluent. But if we want to use it in a higher efficient way, we may make use of engineering machine. Here we show two possible devices:
The first one is Bacterial Board, we can see from DEVICE I.
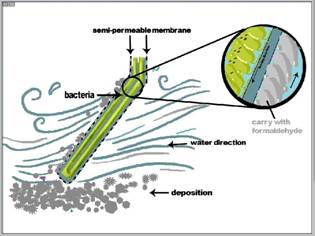
This device is mainly composed by two semi-permeable membranes and a container that contains bacteria, this device is inclined against water direction, and this can limit the influence of deposition in effluent to semi-membrane. Bacteria in container is access to culture jar, it can keep flowing and provide bacteria with successive nutrient and oxygen. It can be used to effluent that has low poison. After setting series of these devices in an area of sewage duct, we can clean formaldehyde efficiently.
The second one is bacterial windmill, we can see from DEVICE II,
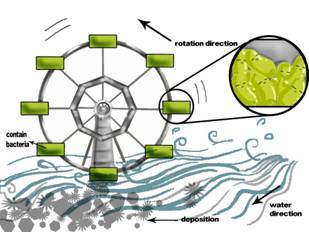
The windmill has a lot of containers, these containers contain bacteria, when the windmill rotates toward water direction, they can contact effluent one by one, and then it can be above the surface of effluent, it provides bacteria with chance to get both formaldehyde and oxygen, bacteria can also avoid being submerged in harmful effluence for long time, and it can also limit the influence of deposition in water. This device can be used in high poisonous effluence.
reference
- [1] Yurimoto H, Hirai R, Matsuno N, et al. HxlR, a member of the DUF24 protein family, is a DNA© binding protein that acts as a positive regulator of the formaldehyde©inducible hxlAB operon in Bacillus subtilis[J]. Molecular microbiology, 2005, 57(2): 511-519.
- [2] Huyen N T T, Eiamphungporn W, Mäder U, et al. Genome© wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol© dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR© family regulator YraB (AdhR)[J]. Molecular microbiology, 2009, 71(4): 876-894.
- [3] Yasueda H, Kawahara Y, Sugimoto S. Bacillus subtilis yckG andyckF Encode Two Key Enzymes of the Ribulose Monophosphate Pathway Used by Methylotrophs, and yckH Is Required for Their Expression[J]. Journal of bacteriology, 1999, 181(23): 7154-7160.
- [4] Ito K, Takahashi M, Yoshimoto T, et al. Cloning and high-level expression of the glutathione-independent formaldehyde dehydrogenase gene from Pseudomonas putida[J]. Journal of bacteriology, 1994, 176(9): 2483-2491.
- [5] Sheng B, Zheng Z, Lv M, et al. Efficient Production of (R)-2-Hydroxy-4-Phenylbutyric Acid by Using a Coupled Reconstructed d-Lactate Dehydrogenase and Formate Dehydrogenase System[J]. PloS one, 2014, 9(8): e104204.
- [6] Khangulov S V, Gladyshev V N, Dismukes G C, et al. Selenium-containing formate dehydrogenase H from Escherichia coli: a molybdopterin enzyme that catalyzes formate oxidation without oxygen transfer[J]. Biochemistry, 1998, 37(10): 3518-3528.
- [7] Winson M K, Camara M, Latifi A, et al. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa[J]. Proceedings of the National Academy of Sciences, 1995, 92(20): 9427-9431.
- [8]Lin L J R, Liao C C, Chen Y R, et al. Induction of membrane permeability in Escherichia coli mediated by lysis protein of the ColE7 operon[J]. FEMS microbiology letters, 2009, 298(1): 85-92.
- [9] Chak K F, Kuo W S, James R. Cloning and characterization of the ColE7 plasmid[J]. Journal of general microbiology, 1991, 137(1): 91-100.
1 Coloration system
We have successfully amplified the P-formaldehyde gene (595bp) from Bacillus subtilis 168 using different annealing temperature(60℃, 58℃, 56℃ corresponding to products 1, 2, 3 respectively).[1]
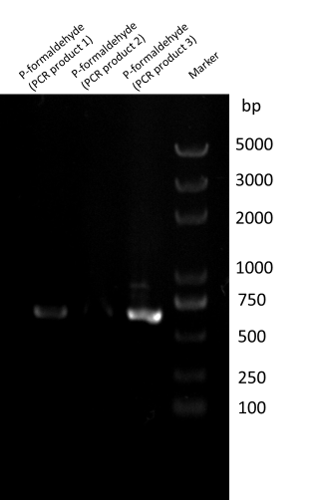
Then we ligate P-formaldehyde with the pSB1C3 backbone using standard assembly to make BBa_K1334002.
In order to prove the function of P-formaldehyde in the system described above, we add a GFP reporter gene to the downstream of it. Thus by detecting relative fluorescence intensity we get to know whether the P-formaldehyde works.[1][2][3]
We transform P-formaldehyde+GFP plasmid into E.coli strain DH5α. The positive clone is screened out and used as experimental group. After cultured to OD600 reaching 1.0 (mid-log phase of E.coli growth curve), formaldehyde is added to broth to 1mM. bacteria samples are collected at different time and their fluorescence intensity and absorbance are measured by microplate reader.
From the following two figures, we can see significant increase in relative fluorescence intensity after formaldehyde stimulation, indicating that our coloration system works well. In addition,the growth condition curve shows that the difference of the fluorescence/absorbance is not caused by the amount of bacteria.
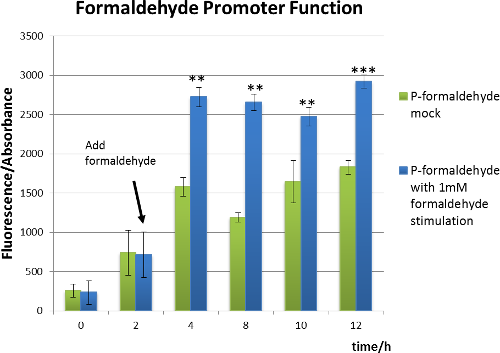
Figure 4 Formaldehyde promoter function chart.
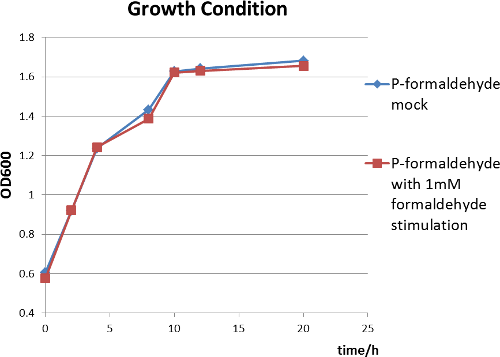
Figure 5 The growth condition of E.coli strain DH5a after formaldehyde stimulation.
2 Street Cleaner System
We construct biobricks: Part: BBa_K1334016, BBa_K1334015 and BBa_K1334017. And we construct a plasmid as following:
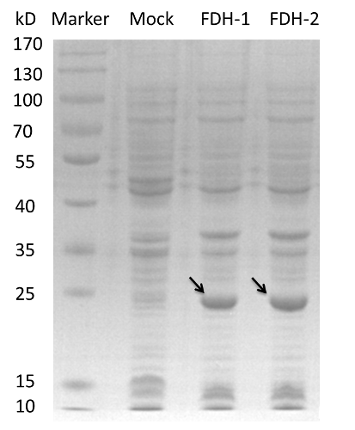
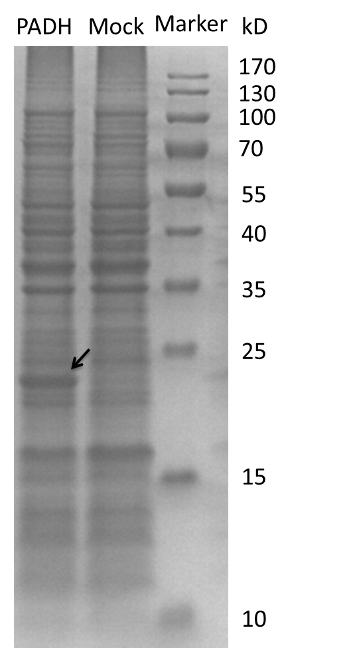
And we transform the plasmids into the expression strain of E.coli. we incubate the E.coli and use the bacteria lysate to run a SDS-PAGE. The FDH from E.coli is about 80kD while we can see a 25kD fragment and the protein PADH is 42.5kD while there is a fragment between 15 and 25kD region. So we need to modify our biobrick to express the two enzymes.[4][5][6]
3 lysis system
(1) Lux system
As we have mentioned in our design, we construct a biobrick to test whether the lux system works or not.
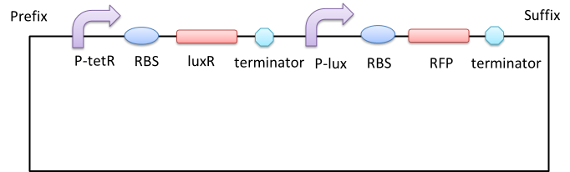
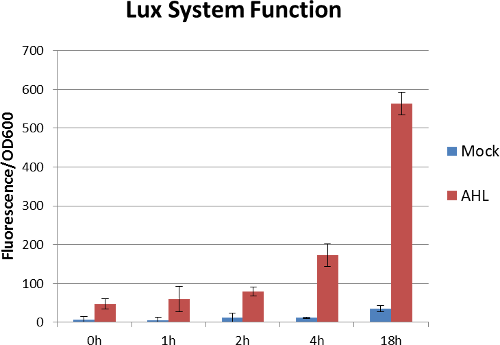
We transform this plasmid into the E.coli strain DH5a. After exogenous AHL (3OC6HSL ,final concentration:100nM)stimulation, we detect the relative fluorescence intensity of RFP. The following figures shows that the lux system works well.[7]
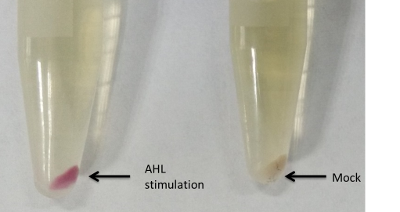
(2) E7 protein
We ligate e7 gene with a commercialized plasmid backbone called pUHE21, which expresses E7 protein when there is IPTG induction.
W3110 is a wild type strain of E.coli that can secret colicin. According to some research, E7 lysis protein can cause the lysis of w3110 strain membrane so that they can release colicin to the extracellular space to kill other E.coli strain (w3110 itself can resist colicin because of the expression of immunoprotein).[8][9]
E.coli strain W3110 with or without E7 lysis gene integrated in plasmid pUHE21 are cultured to OD=0.5 and then induced with or without IPTG. We collected bacteria samples every hour from the fermented liquid and the Optical Density are measured by microplate reader. The results are shown below.
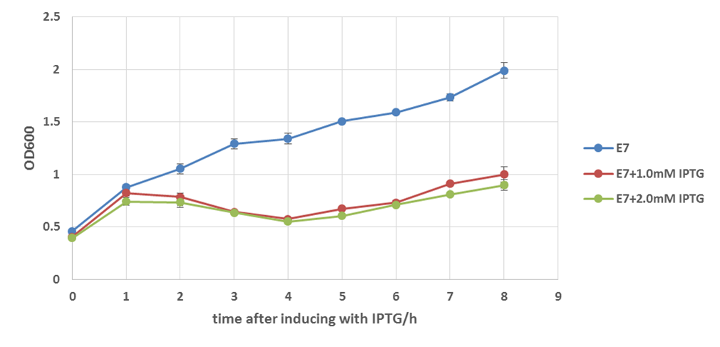
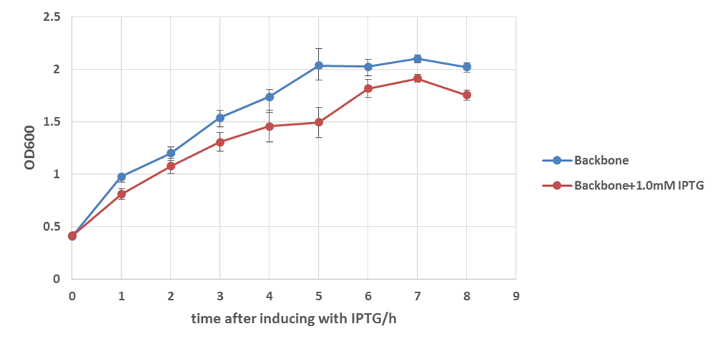
From Figure 13 we get to know that the expression of E7 lysis protein can induce the lysis of colicin-secreted E.coli. Although at the beginning of the induction, the OD of those 3 groups are slightly different with each other because of accidental error, after 2 hours of induction, the differences in OD are too obvious to be considered as the consequence of the error, and after 8 hours induction, the difference even reaches as great as 1 fold.
Besides, Figure 14 can prove that IPTG is not the main cause of the decrease growth rate of E.coli. we know that IPTG itself may do some harm to E.coli, on the other hand, the induction with IPTG greatly increases the expression level of genes downstream pLac ,which may bring additional burden to E.coli as well. So in order to make sure the difference showed in Figure 14 is mainly caused by the lysing function of E7 lysis protein other than IPTG, w3110 without E7 lysis gene on its plasmid (pUHE21, also referred to as backbone) is induced with same concentration of IPTG. The result shows that IPTG can affect the growth rate of E.coli to some degree, but its influence is not strong enough to cause 1 fold difference. Thus we can say that E7 lysis protein is functional to cause the lysing of E.coli and decrease its growth rate.
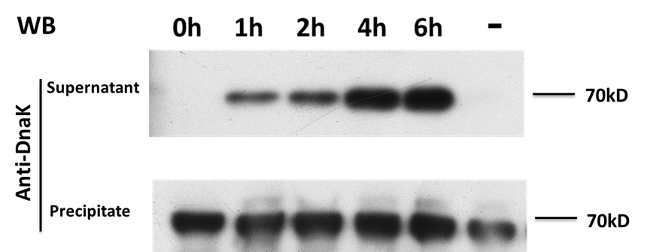
We also use the collected bacteria and the supernatant to do western-blot,using anti-DnaK antibody. (DnaK, or Heat Shock Protein 70 is coded by a house-keeping gene and it cannot go through the cell membrane freely.) We find DnaK in the supernatant largely increases when E7 expresses, which means that the E7 protein do lyse the bacteria.
Reference
- [1] Yurimoto H, Hirai R, Matsuno N, et al. HxlR, a member of the DUF24 protein family, is a DNA-binding protein that acts as a positive regulator of the formaldehyde-inducible hxlAB operon in Bacillus subtilis[J]. Molecular microbiology, 2005, 57(2): 511-519.
- [2] Huyen N T T, Eiamphungporn W, Mäder U, et al. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR)[J]. Molecular microbiology, 2009, 71(4): 876-894.
- [3] Yasueda H, Kawahara Y, Sugimoto S. Bacillus subtilis yckG andyckF Encode Two Key Enzymes of the Ribulose Monophosphate Pathway Used by Methylotrophs, and yckH Is Required for Their Expression[J]. Journal of bacteriology, 1999, 181(23): 7154-7160.
- [4] Sheng B, Zheng Z, Lv M, et al. Efficient Production of (R)-2-Hydroxy-4-Phenylbputyric Acid by Using a Coupled Reconstructed d-Lactate Dehydrogenase and Formate Dehydrogenase System[J]. PloS one, 2014, 9(8): e104204.
- [5] Khangulov S V, Gladyshev V N, Dismukes G C, et al. Selenium-containing formate dehydrogenase H from Escherichia coli: a molybdopterin enzyme that catalyzes formate oxidation without oxygen transfer[J]. Biochemistry, 1998, 37(10): 3518-3528.
- [6] Zinoni F, Birkmann A, Stadtman T C, et al. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli[J]. Proceedings of the National Academy of Sciences, 1986, 83(13): 4650-4654.
- [7] Winson M K, Camara M, Latifi A, et al. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa[J]. Proceedings of the National Academy of Sciences, 1995, 92(20): 9427-9431.
- [8]Lin L J R, Liao C C, Chen Y R, et al. Induction of membrane permeability in Escherichia coli mediated by lysis protein of the ColE7 operon[J]. FEMS microbiology letters, 2009, 298(1): 85-92.
- [9] Chak K F, Kuo W S, James R. Cloning and characterization of the ColE7 plasmid[J]. Journal of general microbiology, 1991, 137(1): 91-100.
If you want to look into details about the functions of our project, please enter the following models:
Formaldehyde promoter model
Objective
To predict the effect of the P- formaldehyde on recombinant plasmid in Escherichia coli, we develop a mathematical model that describes the strength of the P- formaldehyde, and in this model we simulate the procedure of which P- formaldehyde is activated and then expresses the downstream genes, whose expression intensity indicates the strength of P- formaldehyde.
Model Description
1.1The regulation mechanism of formaldehyde operon (HxlAB operon) HxlAB operon encodes two key enzymes in the ribulose monophosphate pathway that are involved in formaldehyde fixation, 3-hexulose-6-phosphate synthase and 6-phospho-3-hexuloisomerase. Expression of the hxlAB operon is induced by the presence of formaldehyde.
HxlR protein can enhance the operon to some extent. The hxlR gene that encodes HxlR protein is located upstream of the hxlAB operon. When the hxlR protein binds BRH1 operating site, the complex can promote the binding of another hxlR protein with BRH2 operating site. On the contrary, binding of hxlR to BRH2 operating site can not promote the binding of BRH1 site. In other words, the binding is in a specified order. Reactions below are involved in the regulation process of HxlAB operon.
\[ hxlR+BRH1\rightarrow hxlR\cdot BRH1(=STR1)\] \[ STR1+BRH2+hxlR\rightarrow 2hxlR\cdot BRH1\cdot BRH2(STR2) \] \[ STR1+STR2+TRP\rightarrow BRH1+BRH2 \]Explanation of the symbols used above: hxlR is for the protein that has enhancing function, BRH1 and BRH2 are for two operating sites, TRP is for the inactive operating sites.
1.2 Transcription efficiency factor \(\eta\)
Based on the equation above we can deduce the following equations.
\[ \frac{[hxlR]\cdot [BRH1]}{[STR1]}=K_{1} \] \[ \frac{[STR1]\cdot [BRH2]\cdot [hxlR]}{[STR2]}=K_{2} \] \[ \frac{[TRP]\cdot [STR1]\cdot [STR2]}{[BRH1]\cdot [BRH2]}=K_{3} \]And the equations of material balance
\[ [hxlR_{t}]=[hxlR]+[STR1]+[STR2]+[BRH2] \] \[ [O_{t}]=[BRH1]+[BRH2]+[TRP] \]Explanation of symbols used above: \( [hxlR_{t}]\) is for the total concentration of the hxlR enhancing protein, \( [O_{t}]\) is for the total concentration of theoperating sites.
Given that there is constitutive instability inside the cell, the aggregate level of operon \( [O_{t}]\) is identical to that of heterologous gene[G]
\[ [O_{t}]=[G]\]The quantity of transcription efficiency factor is defined by the ratio of \( [O]\) and \( [O']\). According to the equation above, we learned that
\[ \eta =\frac{[O]}{[O']}=\frac{\sqrt{[B(A-G)+C]^{2}+4\cdot B\cdot C\cdot G}-[B(A-G)+C]}{2\cdot B\cdot C} \]A, B, C, and G are defined as:
\[ A=[hxlR_{t}] \] \[ B=\frac{[hxlR]^{2}}{K_{1}\cdot K_{2}\cdot K_{3}}\] \[ C=1+\frac{[hxlR]}{K_{1}}+\frac{[hxlR]^{2}}{K_{1}\cdot K_{2}} \] \[ G=[G] \]1.3 mRNA concentration
Supposing that the transcription and the degradation process of mRNA inside the cell are both first order reactions and considering the dilution effect imposed on mRNA by the growthand multiplication of the cells, we learned that
\[ \frac{d[mRNA]}{dt}=\eta \cdot k_{m}\cdot [G]-k_{-m}\cdot [mRNA]-\mu \cdot [mRNA]\]Explanation of symbols used above: \(k_{m}\) is for mRNA transcription rate constant, \(k_{-m}\) for degradation rate constant, \([mRNA]\) for the level of mRNA inside the cell, and for the velocity of cells' growth.
Considering the transcription of mRNA in the cell is basically quasi-stable, so
\[ \frac{d[mRNA]}{dt}=0 \]mRNA level can be derived by the equation above
\[ [mRNA]=\frac{\eta \cdot R_{m} \cdot G}{k_{-m}+\mu} \]1.4The quantity of expression products in the cell
Assuming that the expression of products as well as the degradation of mRNA is a first order reaction of the quantity of mRNA inside the cell, and considering the dilution effect imposed on the products by the growth and multiplication of the cells, we learned that
\[ \frac{d[P]}{dt}=k_{p}\cdot [mRNA]-k_{-p}\cdot [P]-\mu \cdot [P] \]Given that the expression of the products inside the cell is basically stable, thus:
\[ \frac{d[P]}{dt}=0 \]According to the equation above, we can solve for the level of cell products
\[ [P]=\frac{k_{p}\cdot [mRNA]}{k_{-p}+\mu} \] \[ [P]=\frac{|\sqrt{(B(A-G)+C)^2+4\cdot B\cdot C\cdot G}-[B(A-G)+C]|\cdot k_{m}\cdot k_{p}}{2\cdot B\cdot(k_{-m}+\mu )\cdot(k_{-p}+\mu )\cdot\rho } \]1.5 The concentrationof products during the fermentation process
According to the equation above, we can figure out the concentration of products in the fermentation broth, which can be described as
\[C_{p}=\frac{|\sqrt{(B(A-G)+C)^2+4\cdot B\cdot C\cdot G}-[B(A-G)+C]|\cdot k_{m}\cdot k_{p}\cdot x}{2\cdot B\cdot(k_{-m}+\mu )\cdot(k_{-p}+\mu )\cdot\rho }\]Explanation of symbols used above: C is for the level of the products during fermentation process (\(\mu\)/L), x is for the level of cells in the fermentation broth (g/L), and I is for the density of cells (g/L)
From the result given above we can infer that only when the promoter is activated, can hxlR accumulate constantly. Besides, the probability of its binding with BRH1 and both BRH1 and BRG2 will increase, and as a result, the metabolites in bacteria cells will increase.
Result and Analysis
In our actual experiments, we use RFP as downstream expression product (final metabolite of cells). After stimulating the growing bacteria with formaldehyde, we measure the fluorescence intensity of bacteria every once in a while, the results (the greater the fluorescence intensity, the greater the expression of RFP) are shown in Figure 1.
As can be seen from Figure 1, after the stimulation of formaldehyde, the fluorescence intensity increases significantly, indicating that the model predictions matches the result of experiment, which also proves the correctness of our modeling.
Reference
[1]A first course in mathematical modeling / Frank R. Giordano ... [et al.] China Machine Press, 2009. [2] A Guide to MATLAB : For Beginners and Experienced Users. Cambridge Cambridge University Press, 2012.iGEM Medals for Environment Teams
Requirements for a Bronze Medal:
Register the team, have a great summer, and plan to have fun at the Giant Jamboree.
Successfully complete and submit this iGEM 2014 Judging form.
Create and share a Description of the team's project using the iGEM wiki and the team's parts using the Registry of Standard Biological Parts.
Plan to present a Poster and Talk at the iGEM Jamboree.
The description of each project must clearly attribute work done by the students and distinguish it from work done by others, including hosting labs, advisors, instructors, sponsors, professional website designers, artists, and commercial services. Please see the iGEM 2011 Imperial College Acknowledgements page for an example. Link to page on your team's wiki: Attribution
Document at least one new standard BioBrick Part or Device used in your project/central to your project and submit this part to the iGEM Registry (submissions must adhere to the iGEM Registry guidelines). Please note you must submit this new part to the iGEM Parts Registry. Please see the Registry help page on adding new parts. A new application and/or outstanding documentation (quantitative data showing the Part's/ Device's function) of a previously existing BioBrick part also counts. Please see the Registry help page on how to document your contributions. To fulfill this criterion, you will also need to submit the part with its original part name to the Registry, following the submission guidelines.
Part Number(s):
BBa_K1334002 [Received, Accepted] BBa_K1334020 [Received, Accepted] BBa_K1334000 [Received, Accepted] BBa_K1334001 [Received, Accepted] BBa_K1334003 [Received, Accepted] BBa_K1334004 [Received, Accepted] BBa_K1334005 [Received, Accepted] BBa_K1334006 [Received, Accepted] BBa_K1334007 [Received, Accepted] BBa_K1334008 [Received, Accepted] BBa_K1334015 [Received, Accepted] BBa_K1334016 [Received, Accepted] BBa_K1334017 [Received, Accepted] BBa_K1334018 [Received, Accepted] BBa_K1334019 [Received, Accepted] BBa_K1334020 [Received, Accepted] BBa_K1334021 [Received, Accepted] BBa_K1334022 [Received, Accepted] BBa_K1334024 [Received, Accepted]
Additional Requirements for a Silver Medal:
Experimentally validate that at least one new BioBrick Part or Device of your own design and construction works as expected.
Part Number(s):
BBa_K1334002 [Received, Accepted] BBa_K1334020 [Received, Accepted]
Document the characterization of this part in the Main Page section of that Part's/Device's Registry entry. (the same linkage as above)
Submit this new part to the iGEM Parts Registry (submissions must adhere to the iGEM Registry guidelines)
Part Number(s):
BBa_K1334002 [Received, Accepted] BBa_K1334020 [Received, Accepted]
iGEM projects involve important questions beyond the bench, for example relating to (but not limited to) ethics, sustainability, social justice, safety, security, or intellectual property rights. Articulate at least one question encountered by your team, and describe how your team considered the(se) question(s) within your project. Include attributions to all experts and stakeholders consulted. Link to page on your team's wiki: Research: Formaldehyde in cars
Additional Requirements for a Gold Medal: (one OR more)
Improve the function OR characterization of an existing BioBrick Part or Device (created by another team or your own institution in a previous year), enter this information in the Registry. Please see the Registry help page on how to document a contribution to an existing part.
Part Number(s):
We contribute to the part BBa_K117000.
Help any registered iGEM team from another school or institution by, for example, characterizing a part, debugging a construct, or modeling or simulating their system. Link to page on your team's wiki: Helpling Other Teams
iGEM projects involve important questions beyond the bench, for example relating to (but not limited to) ethics, sustainability, social justice, safety, security, or intellectual property rights. Describe an approach that your team used to address at least one of these questions. Evaluate your approach, including whether it allowed you to answer your question(s), how it influenced the team's scientific project, and how it might be adapted for others to use (within and beyond iGEM). We encourage thoughtful and creative approaches, and those that draw on past Policy & Practice (formerly Human Practices) activities. Link to page on your team's wiki: Research: Formaldehyde in cars
Research: Formaldehyde in cars
Concentrating On the Safety, Leading Our Design
Before we started our project "formaldehyde terminator", we aimed at detecting the existence of formaldehyde in the houses, but as time went by, we realized that high formaldehyde level is a more serious problem in cars. "Although we can tell whether the level of formaldehyde is high or not, we have almost no methods to eliminate formaldehyde in cars quickly." So we improved our design by adding Street Cleaner System, so we could not only detect formaldehyde but also remove it continuously.
Introduction
As we know, formaldehyde exists not only in factories and some extreme environment but also in our daily life, but at the same time we often ignore the formaldehyde in some basic area around us, we aim nat finding what we used to ignore, and finally we have done the research of detecting formaldehyde in cars. In this program, we use formaldehyde kits to help both teachers and our classmates to detect formaldehyde level in their cars. 34 cars have been tested and finally almost one third of these cars’ air-formaldehyde condition is excessive. Besides these, we have also done some other analysis about different factors’ influence on formaldehyde level.
You can download the detail information from here.


Analysis
After we have collected series of data, we did a lot of analysis. For the reason that our data are influenced by different factors, we set groups of two-variables analysis (other factors are ignored when analyzing one group). Here are our data:

Table 1.1 The rate of excessive cars and compliance cars
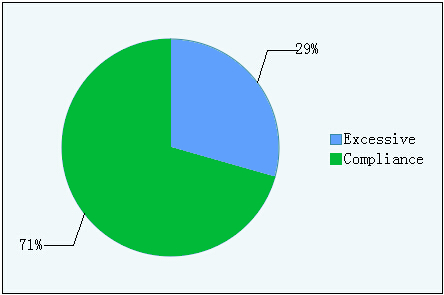
Figure1.1 The rate of excessive cars and compliance cars
The cars which have excessive level of formaldehyde are about one third to all cars tested.
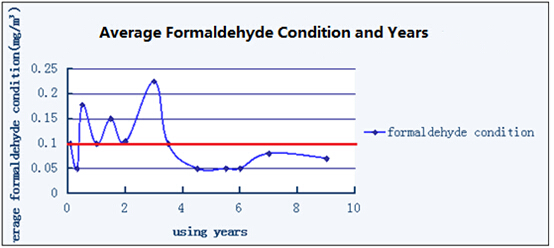
Figure1.2 Time and cars Red line stands for Cordon - It is the content of formaldehyde higher than 0.1mg/m3 that is excessive.
Using years is related to level of formaldehyde in cars closely, through the graph we can find that there exists a clear boundary between the excessive group and the normal group, new cars are of high possibility to have excessive level of formaldehyde while old cars may remain normal. (For the reason that quantity of our data is limited and unknown factors may also influence the research, we can only get the rough conclusion)
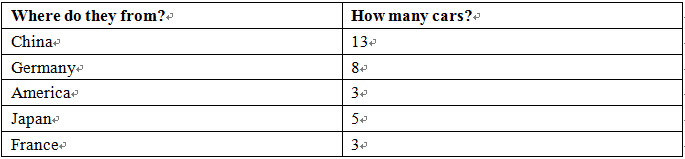
Table 1.2 Countries and cars
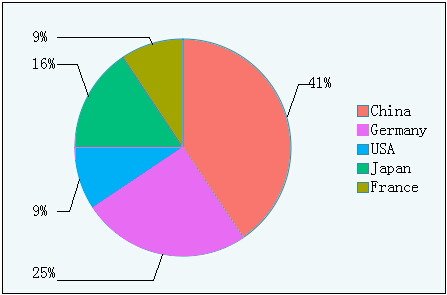
Figure1.3 Rate of cars produced from different countries
Besides the quantity, we also did the analysis of the relationship between countries and average formaldehyde level of cars.
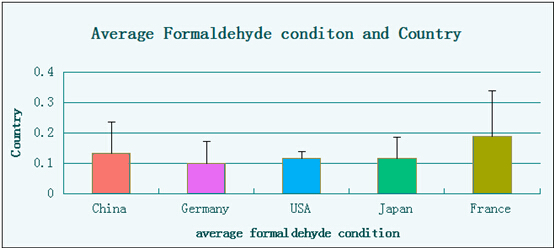
Figure1.4 Average level of formaldehyde in different groups.
From this graph we can know clearly that cars from different countries have diversity in the level of formaldehyde, so we may think it over before deciding which one to buy.
Conclusion
From this program we have finished, we can find that the basic vehicle - cars may have higher concentration of formaldehyde than we thought before (For the reason that nearly 30 percent of cars may have excessive level of formaldehyde). It alerts us that we should focus our attention on what we may ignore before, even the basic one around us. Through this program, teachers and our classmates have got access to our iGEM project, almost all of them participated in it happily and finally found what was ignored before. This is not only good for communication between teachers and students, but also can help more people to realize environmental problems in our daily life. We feel sorry for that kits can only detect but not remove formaldehyde at the same time, if our iGEM project could come true, we can solve this problem easily. More than one teacher have shown their interest in our project of using E. coli to detect and remove formaldehyde at the same time, we will work harder and try all we can to make our dream come true!
Helping other teams
Giving iGEM14_NUDT_CHINA A Biobrick We Constructed
We give iGEM14_NUDT_CHINA a new biobrick we constructed this year. The part number is BBa_K1334030. They used this part in their project and test its function. Thanks for their acknowledgement in theri Experiment Support.
Helping iGEM14_HUST-INNOVATORS Constructing Their Biobrick
In September in 2014, after we finished our project probably and went to the last step, we helped iGEM14_HUST-INNOVATORS construct a biobrick BBa_K1551000.
The biobrick: BBa_K1551000.
The reference page of iGEM14_HUST-INNOVATORS (Please click Team-intro.-Special Thanks)
iGEM14_HUST-INNOVATORS is a team which consists of intelligent guys whose major are not biology-relevant. They made a piece of fascinating software and found a biological company to synthesize a part in pUC57 backbone to generate delta-5 fatty acid Elongace while they had difficulties in transforming the part into pSB1C3. We changed the backbone for them and helped them by adding the part to the registry.
Figure2.1 Detail information of plasmid pUC57
After days of hard working, we finished this project successfully, here shows the final result of electrophoresis.
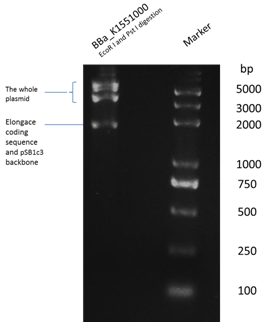
Figure 2.2 We digest Bba_K1551000 with restriction enzymes EcoR I and PstI . The Elongace coding sequence (with prfix and suffix)is 2064 bp while the pSB1C3 is 2070 bp so that there is only one bar near the 2000 bp region, which shows that we change the plasmid backbone successfully.
Participating in other teams' human practice
In June, we got an e-mail from the team Virginia, from this e-mail we knew that they were preparing for a global survey about synthetic biology, we participated in their survey actively and finally helped them to collect nearly 50 surveys, they provided us with a nice badge and thanked us for helping.
Email from Virginia:

Besides, we also helped XMU-China to finish their Newsletters, we were very happy to see their Newsletter every week.
Email from XMU-China:

Helping other teams after CCiC:
Besides the information mentioned above, after the CCiC(Central China iGEMer's Consortium), we amplified our communication with other teams such as HZAU-China, NJAU-China, NUDT-China and so on, we also provided them with help, here is the brief introduction:
Before the CCiC, we convened a preparation meeting in WHU-China, and helped HZAU-China to formed CCiC. We also exchanged some parts with HZAU-China, such as dCas9 device(BBa_K1081000)(we constructed this part last year), and luxI generator: BBa_K1334030(we constructed the part this year)
Special thanks from HZAU-China
After communicating with NJAU-China, we knew that their program is related to dCas9 device, which we had constructed the last year, so we also provided them with BBa_K1081000.
After communicating with NUDT-China, we knew that both of our projects would use N-(3-oxododecanoyl) homoserine lactone (AHL for Las system), at that time we had gotten AHL, so we sent AHL to them, besides, we provided BBa_K1334030 to them. Similar situation happened to BIT, which team had also received AHL from us.
Popularization
During our process in finishing the project, we had also tried to introduce iGEM to more people around. To achieve our goal, we had hosted series of wonderful activities.
In May, at the very beginning of our project, we hosted Scrawl Competition in Microorganism, in this competition, every participant would get a culture plate and E. coli which contained GFP, they could use inoculating loop to draw what they like on the culture plate.


In September, we utilized the chance of welcoming the fresh to our university to advertise iGEM, in order to bring them knowledge about iGEM as well as happiness, we hosted series of activities. When the fresh come to university to register, they will get a bag which contains books about the university, card they will use in daily life and other guide that can help them to adapt the university. We added our poster and an exquisite badge into each bag, they were very happy when getting gifts. In addition, we also hosted a meeting to welcome them and advertise iGEM, our header Sun Haoyuan gave them a wonderful speech and told them a lot about his experience in university and iGEM.



Further than advertising to students around us, we have taken other ways to expand our influence. One way is through the internet, we have set up our own Xinlang Weibo, public pages on Renren and QQ groups, we keep telling the public about iGEM and the process of our experiments. Till now on ,we have a lot of fans, and they can learn numerous knowledge from us. Another way is participating in competition hosted by our school, in the early October, we took our program to Creation Competition in our school, and we showed our idea to many teachers in different majors and finally got a great score.
Communication
We have communicated with a lot of universities and organizations. First of all we will introduce you CCiC here. CCiC is Central China iGEMer's Consortium. In the early April, HZAU-China, HUST-China and WHU-China ,WHU-Pharm worked together to prepare for this great meeting, and CCiC finally began at 23rd August successfully in Huazhong Agriculture University. Almost 20 universities from all over the China came to Wuhan to attend the meeting. We made friends with students from these universities and at the same time we communicated and exchanged our great ideas with each other.



Besides CCiC, we also communicated with a lot of teams. Here is a list that notes the wonderful experience:
Our leader went to Jilin University and had a nice communication with them, besides exchanged our ideas, he invited Jilin_China to join in CCiC and have a visit to Wuhan University.

Our advisor Jia Hangxing was invited by friends from SCU to visit SiChuan University and they had a wonderful talk in May.

Friends from OUC-China, SCUT and SCAU-China visited Wuhan University after CCiC. Students from WHU-China and WHU-Pharm welcomed them and guided them a wonderful trip.

Members

Hello everyone, Here is the team of WHU-China! Team WHU-China 2014 was born at the foot of the mount LuoJia, November 2013. The team is mainly formed by sophomore and junior of the college of life sciences, Wuhan University. Our team has a great tradition and this year is the third time to join in the competition. In our warm small group, both sophomore and junior work together. We hope that we can learn more knowledge of synthetic biology and get access to more experimental skills by ourselves during the competition. What is more important is that we can make a contribution to the society. Let us join hands to swim in the vast ocean of synthetic biology and find treasures one by one!

Haoyuan Sun, as the captain of the team, is devoted himself into the completion. From the establishment of the project, to undertake the whole experiment guideline, he is just like a glaring star in the night to guide us where to go ahead. In the iGEM team, he not only uses his knowledge to solve the problems, but also encourages us by his humor to get through the difficulties. Besides the iGEM, he is skilled in playing ball games and telling jokes.

Hi, I am Zheng Sun, In the team of iGEM, everyone can find his/her own place, and iGEM has different meanings to different people. As for me, iGEM provides me with a great chance to explore the field of synthetic biology and the way to go into the society to touch different people and handle series of things I have never met before. Thanks iGEM for giving me such a chance and I believe this wonderful experience can leave me the beautiful memory and treasure in my life.

Here is Yun Xue, this is my first year in iGEM. I joined iGEM because I'm fond of synthetic biology and I'm skilled in the mathematical modeling, genetics and molecular biology. And I would like to pursue these interests in future and do research about mathematical modeling of biological systems. I enjoy sport activities such as badminton, tennis and basketball. I am lookng forward to solving problems and getting good achievements.

Qiushuang Wu: the smart girl provided us with the original idea for our project this year in the brainstorming. As the housekeeper in our lab, she kept the lab organized when doing experiments. And she herself did a lot of experiments, involving all parts in our project. As some people's comment, she was always in the lab or on the way to the lab. She usually says if love, please love deeply. She loves biology very much, and will devote to her true love all her life.

Long Yu, coming from Hubei, China, majors in biology in College of Life Sciences, Wuhan University. In his second year, he entered into the lab of Prof. Chen, whose field is microbiology. During those times, he was mainly devoted to reveal a new kind of mechanism by which genetic materials were transferred between Bacilus subtilis other than three traditional horizontal gene transfer (HGT) ways. In his third year, he participated in this iGEM team, and devoted much time into it, so granted, he learned a lot. But it is a pity that he could not keep it up. Now in his fourth year in college, he was prepared to go to Monash University in Australia, conducting researches about Tfh cells.

My name is Han Dong. I have been studying in Wuhan University for more than 2 years, majoring in biology. I was born in a coastal city named Qingdao, where I grew up with the sea, thus I like seafood, beach and sea breeze. Sea makes me open-minded, willing and have broad interests like swimming. I also enjoy reading and drawing, so I am the main graphic designer in our team. I believe that painting will help people to learn the world better by watching it carefully, and it is the same with biology. That is why I join the team , I want to show our idea with my pen.

I am Teng Li, a girl who loves reading and wants to make everyday in my life meaningful. I regard being an iGEMer a happy thing that deserves my devotion. All the iGEMers in our team are my friends and I do enjoy performing experiments and exchanging ideas with them. It is in iGEM that I learned how to cooperate, how to think and the basic skills for doing research of synthetic biology.

Hello! I am Zhengyu Luo, a senior student major in math and I am the computer engineer of the team in charge of the construction of the webpage. I like art and design and I am full of energy and prepared for any kind of challenges. Synthetic biology is a very hot subject these days so I joined WHU-China to learn something about it. I believe it must be a very interesting experience.

Hey guys! I'm Li Yuwenbin from Wuhan University, a team member of Whu-China. My job in our team mainly focuses on proving the function of E7 lysis protein as well as integrating it into our formaldehyde-terminator system, so that those functional enzymes can be secreted out into extracellular space to degrade formaldehyde more efficiently. Besides biological research, I have a strong passion for various kinds of sports, like cycling, badminton and even x-game. I also have a special taste for music, poems and animation. iGEM jamboree has a lot of fun and it's so great to meet new friends and exchange ideas with each other in the competition. See you guys at MIT^ ^!

Hi there, this is Tianxing Zhai, a student of College of Life Sciences at Wuhan University. As a team member of WHU-China, I was responsible for the functional verification of formaldehyde dehydrogenase (spectrophotometry), some molecular cloning, and other non-experimental activities (e.g. ordering uniforms and badges). I am interested in making science more interesting and more widespread. If you and I are like-minded, please contact me. My Email is txzhai@whu.edu.cn.

Hi, I'm Bei Liu. I was in the lab of proteomics in Wuhan University. I did some experiments of biochemistry in the early stage. I also helped Haoyuan in organizing the team and putting up some ideas. Besides synthetic biology, I'm also interested in proteomics and neurogenesis. Now I'm doing an internship in UT Southwestern medical center in Dallas. It is really cool to participate in the giant jamboree in Boston, because Dallas is just like a large valley, while Boston is the true Big City. I hope I could spend some time visiting the city in the spare time during the jamboree.

I am Yao Chen, I still remember the first time I knew something nabout iGEM when I was a freshman, my classmate joined this team whereas I could only listen to his proud talking. Until last November I participated in the iGEM team for my dream. We experienced the hard Saturday night brainstorming and staying up late for presentation of our project and finally our sub-team project became the only project in the team. First I even could not understand what we would do but thanks for the team members, I made it clear ultimately. To retrospect the last 10 months we went through the hardness and success which enrich my experience, broaden my horizon and give me friends. Now I am placid to write the story between iGEM and me and appreciate all the people helped us.

I am Shuohui Liu. During the four years in university, one could choose to sit around and achieve nothing. Or one could treasure the time to do something meaningful. The most meaningful thing for me is to join the iGEM. In the iGEM, one could do researches that are really special and interesting. One could also make friends and work in a team efficiently and comfortably. What's more, one could enjoy the best time in the life. In other words, the one in the iGEM is the most fortunate one in the word. Absolutely, I'm a guy like that.

Zhengwei Yuan| Senior in Biological Sciences Zhengwei is responsible for the separation of FDH (Formate dehydrogenase) gene from Candida boidinii which was bought from China Center of Industrial Culture Collection (CICC) and he work so hard with our team leader that they stay up late for three consecutive days to do the experiment. The main driving force of their diligence is the love for the iGEM and members in our team!

I joined iGEM-team at my second year of colleague. I chose life-science and wished to learn how to design a new life by myself. iGEM gives me the chance to know more and do more. Ideas will never appear suddenly - after the brain storm and layers of screening we got the amazing idea finally and had a colorful life in the lab for several mouths. I joined the human practice group at the last few weeks and did some help. Thanks to my teammates and teachers, we finished it quite well. I love dancing and reading novels. My favorite novel is Harry Potter and I don't think it is childish. Life is colorful, both in the lab and outside.

My name is Lei He coming from Wuhan University school of pharmaceutical science. Because of my enthusiasm and teachers' help, I have got a rich experience of experiment. But being an iGEMer of a great team is the most exciting part.

NAME Shuang Liu BIRTHDAY 1994/01/04 CITIZENSHIP the People's Republic of China NATIVE PLACE Shandong Province HEIGHT 5 feet 10 inches WEIGHT 158 lb BLOOD TYPE A ALLERGY NONE RELIGION ATHEISM

Hi I'm Chen Xi or you can call me Fred. As a senior year student, I'm grateful to be a member of this excellent miracle-making team. My work in this project involves source gathering, plasmids design. I am so glad that our idea and the outcome can now have a chance to be presented to the world and let people know how the bioengineering can create a formaldehyde toxicity free world.

My name is Ji Yang. I am a senior student in College of life science, Wuhan University. It's a great honor to be a part of the 2014-WHU-China team. In the team, my responsibility is to assist Luo Zhengyu to construct our web page and collect some necessary text material. We would use some fancy CSS and JavaScript techniques to make our page more attractive and more creative. I wish our work could present you a clear and insight web page.

Qi Wang| Senior in Biology science Qi is really a shy and introverted girl. She does not like to talk with other members very much, but she is willing to be a good listener. A good team needs not only someone who is eloquent enough to keep us together, but someone who dedicated himself silently like Qi. And she is very serious with her work. She worked in the lab for our project in the summer vacation.

Hi! I am Man Li from WHU-China. I met iGEM one year ago and I am totally into it. In my mind, iGEM is not a competition merely but a club. In this club, students who have the same interests in genetically engineering work together. We set our goals, and achieve some ridiculous ideas. As for myself, I am an outgoing girl. For me, research in biology is one part of my life. In my mind, the quality of life is equally important to the achievement I make in biology. I play violin in my spare times, and enjoy the peaceful afternoons. In WHU-China, I am the one who mobilize the atmospheres in our laboratory and cheer others up.

I'm Rui Bai, a senior student majored in life science, I'm so lucky to be a part of our awesome WHU-iGEM. Before entering the team, I have participated in a molecular biology lab for a year. Because of curiosity, I joined in our team to broaden knowledge of synthetic biology and share ideas with coworkers, which we called brainstorming. Now I think it has been one of the most correct choices which I have made in my college. What's more, I am good at organizing activities for heading up the Student Union. And I also have an appetite for drawing and basketball.

Xi Tu| Senior in Biological Sciences Xi joined the team, wondering how synthetic biology works. He takes part in discuss the idea and good at primer design.
Instructors

Zhaoning Wang

Tong Liang

Hangxing Jia
Advisors

Prof. Zhixiong Xie

Prof. Yu Chen
Attributions
- Prof. Lin Guo from the College of Life Sciences in Wuhan University provided us with lab and equipment to finish parts of our project.
- Prof. Fang Peng from China Center for Type Culture Collection helped us with the culture medium's making and giving information of culturing and separating bacteria.
- Prof. Yin Lei from the College of Life Sciences in Wuhan University helped us to finish the project of detecting formaldehyde in cars, he provided us with formaldehyde electronics test device.
- Friends from Huazhong Agriculture University provided us with the part BBa_J13002, and they also inspired us and entertained us very well during the CCiC.
- Mr. Jiawei Huang, Yan Zeng helped us during the whole process of experiment. They provided us with a lot of new ideas and new methods.
- Mr. Yang Liu helped us to find our way in finishing school internal procedures and getting visa.
- Teachers from the College of Life Sciences in Wuhan University helped us to finish our program of detecting formaldehyde in cars, they participated in it actively and introduced it to their friends. Also thanks our classmates to join in this program.
- Yunyun An, a very clever girl help us with our daily experiments as a technician for several days when we are busy.
- 16 teams have participated in this meetup CCiC. Without them, the first Central China iGEMers' Meetup couldn't be such a happy, exciting and wonderful meetup. These 16 teams are: OUC_China, SCUT, CAU_China, WHU_China, NJU-QIBEBT, SICAU, Jilin-China, AHUT_China, Peking, LZU-China, SCAU_China, BNU-China, WHU-Pharm, HUST-China, BIT, NUDT_China, SICAU.
Overview:Welcome to our Safety Page. We will first answer the safety questions asked by iGEM headquarters briefly, and then discuss safety issues associated with our project in detail..
Timeline
2013
Nov. - Dec., Team building
2014
Jan. - Feb., Knowledge preparing. Looked into iGEM and previous teams’ wiki.
Mar Brainstorming
1st, Put forward our idea to reduce formaldehyde
8th, the idea was made our project purpose. Decided to start with acid promoter
15th, acid promoter plan failed. Start to focus on formaldehyde promoter.
19th, first campus weekly thesis defense.
Apr., Idea building.
10th, last Campus Thesis Defense.
20th, internal adjustment. Established the group framework. Group members increased to 30.
25th, with primers, managed to generate qualified promoters
May, Instructor assessment and suggestions.
3rd, lab cleaning. “Assembly Line” mode was introduced. Timing of preliminary stage.
4th, the result of the identification of the single colony which was positive.
Jun. ~ Oct., Experiment and Human Practice
Jun. 30th, Group meeting. T-easy plasmid was introduced.
Aug. 15th, Group meeting.
Aug. 18th, Group members set off to HZAU for investigation and study.
Aug. 20th, Debug of Human Practice. Team networking.
Aug. 26th, Attended CCiC held in Huazhong Agricultural University.
Sept. 4th, Decided group Logo. First sketch of our poster.
Sept. 6th, Got first result to prove formaldehyde promoter functional.
Sept. 10th, Mathematical Modeling preparation.
Sept. 20th, PowerPoint materials preparation.
Sept. 28th, Biobrick shipping.
Sept. 30th, Another biobrick shipping.
Oct. 1st, Assisted HUST-Innovators in constructing a biobrick
Oct. 4th, Prepare for mathematical modeling and establishment of website
Multi-Assembly-Line Journal:
Promoter
Jun. ~ Sept., Pre-experiment for the construction and function test of hxlR operon.
Aug. 20th, hxlR-GFP completed.
Sept., Got a passable result with a small amount of leakage.
Lux Systerm
Sept. 26th, Construction complete.
Oct. 1st, Function test showed a positive result
.FDH, PADH
Jul. ~ Aug., Enzyme DNA extracting and processing.
E7
Aug. 20th, Lysing function test. Western blot processing and OD600 observing.
Methods
Function Test of E7 Lysis system
The construction of E7-pUHE21 contained strain w3110
BBa_K117000 which contains E7 lysis gene is found out from iGEM 2014 registry and primers are design to PCR the target gene(the sequence of the primers are shown below) BBa_B0034 is contained in the upstream primer, which is a strong RBS.The plasmid pUHE21 is a commercialized one that can be extracted from a modified strain of E.coli(part of its sequence is shown below). EcoRI and PstI are used to cut both the PCR product and the pUHE21 backbone. After purification and ligation with T4 ligase, the reconstructed E7-pUHE21 plasmid is transformed into competent E.coli strain w3110. The positive clone with right DNA sequence inserted is screened out and referred to as strain E7
Sequence of upstream primer with strong RBS (BBa_B0034): CGGAATTCTTCTAGAGAAAGAGGAGAAATACTAGATGAAAAAAATAACAGGGATTAT
Sequence of downstream primer: AACTGCAGGCGGCCGCTACTAGTATTACTGCGTTTCCACTCCG
The construction of pUHE21 contained strain w3110
In order to make a negative control of this experiment, pUHE21 is transformed into competent E.coli strain w3110 and the positive clone is screened out, referred to as strain BB(short for backbone).
IPTG induction and OD measurement
Inoculate 3 Erlenmeyer flasks (referred to as E7-0, E7-1 and E7-2 separately) containing 100ml LB medium with 1ml of saturated overnight strain E7 culture, and at the same time inoculate 2 Erlenmeyer flasks (referred to as BB-0 and BB-1 separately) containing 100ml LB medium with 1ml of saturated overnight strain BB culture. All of the broth are cultured to OD=0.5, then IPTG is added to E7-1, E7-2 and BB-1 to make their IPTG concentration reach 1.0mM, 2.0mM and 1.0mM, respectively. After induction those 5 Erlenmeyer flasks are cultured for another 8 hours, during which 3 samples are taken into a 96-well plates from each flask every hours and their OD are measured with microplate reader.
Western blot to further prove the lysis effect of E7
Besides OD measurement, we also do Western blot. Taken from each Erlenmeyer flasks at 0, 1, 2, 4, 6 hours after induction, those samples are centrifuged ,the supernatant and sediment are used to run a SDS-PAGE, then the antibody of DnaK(which is an endogenous protein expressed constitutively in E.coli) is used to conduct a western blot.
Function Test of The Enzymes
To test whether formaldehyde dehydrogenase (PADH) work or not, we developed this system. In this system, when PADH oxidize formaldehyde, electrons lost by formaldehyde are transferred to nicotinamide-adenine dinucleotide (NAD+). Gaining electrons, the NAD+ are reduced to reduced form of nicotinamide-adenine dinucleotide (NADH). By measuring the absorbance of NADH under light of 340nm, we can know the concentration of NADH, and finally know the activity of PADH.
Culture
Experimental group (E.coli with PADH gene) and control group (E.coli without PADH gene) are both cultured under 37℃, overnight
Lysis
Collect cells by centrifugation, and wash them with Tris-HCl (PH 7.5) twice
Resuspend cells with Tris-HCl (PH 7.5)
Lyse them by ultrasonic cell disrupter: 500w, work 5s, interval 7s, repeat dozens of times until the solute becomes clear.
Collect supernatant by cryogenic centrifugal (4℃, 12 000 r/min , 20min)
Enzyme activity assay
Prepare the reaction system in colorimetric tube: 1.5ml Tris-HCl (PH 8.0), 0.5ml NAD+ (6mM), 0.5ml formaldehyde solution (12mM), 2.5ml total
Incubate under 50℃ for 10min, measure the absorbance from time to time until stabilized
Add 0.5ml supernatant (experimental group) or Tris-HCl buffer (control group) to the reaction system, and continue incubating
Measure the absorbance under 340nm wavelength of light, after 5min, 10min, 15min, 20min respectively


